Motivations
The most important factor in good machine learning systems is the quality of data. A model’s performance lives or dies by the data used to train it.
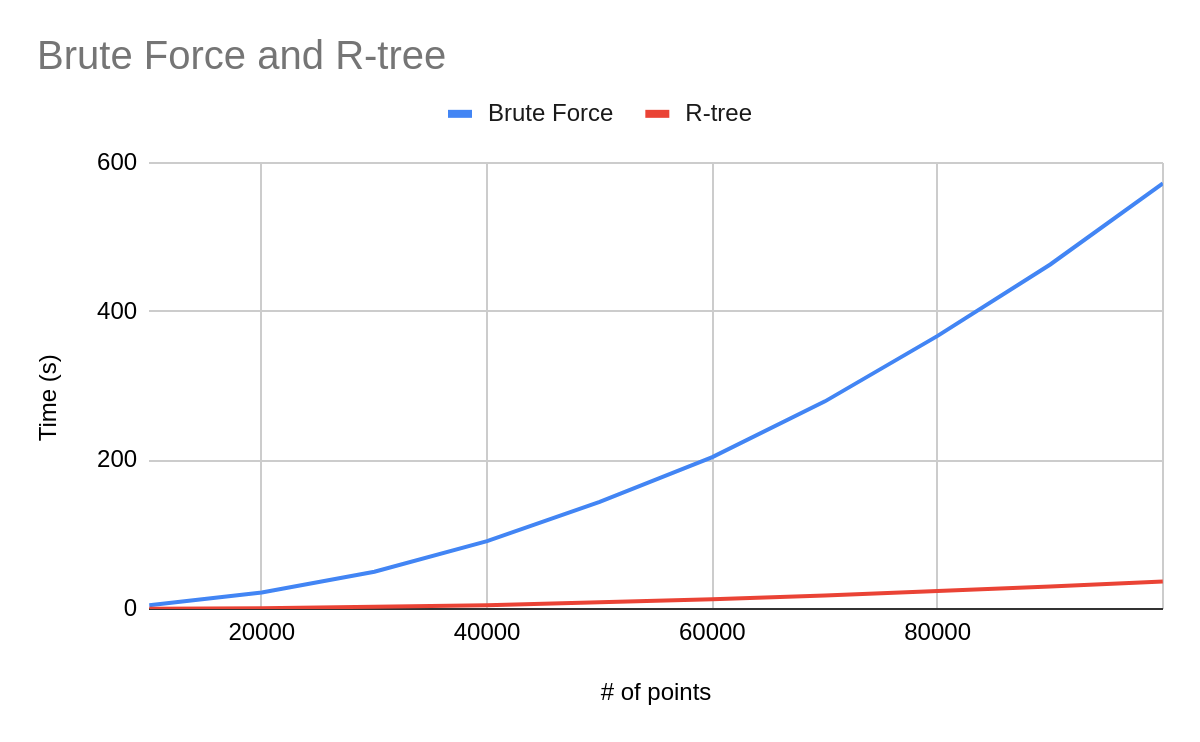
Unfortunately, for the longest time I did not rigorously analyze my model or even input data. My typical approach is to first view the training loss graph. If the accuracy and loss are acceptable, I apply the model on unseen data. Then I look at the F1 scores and confusion matrix to evaluate the performance.
If the metrics are good then I can call it a day. However, what if the model performs horribly? What is the failure mode of the model? Which training data is contributing to problem, and which data are hard to train on? We can easily see how lackluster the aforementioned numbers are when it comes to answering these questions.
Introducing FiftyOne
What if there are methods and tools to help us answer these questions? For those working in computer vision, that is what tools like FiftyOne is built for.
FiftyOne provides the tools for visualizing, analyzing and cleaning training data. It comes with building blocks for optimizing the dataset analysis pipeline.
Some things that FiftyOne can do are:
- visualizing complex data labels
- identifying failure modes
- find annotation mistakes
- -evaluate models
- -explore scenario of interests
To make these tasks easy, FiftyOne has powerful graphical tools that are easy to use. These tools can be easily integrated with things like Jupyter notebooks to make exploratory data analysis more interactive.
Embedding Visualization
Because FiftyOne is so extensive, I will only cover the data visualization tools that are offered.
As stated earlier, staring at aggregate metrics hoping to find ways to improve a model’s performance is a foolish errand. A much better way is to visualize the data in a low-dimensional embedding space. This is a powerful workflow that reveals pattern and clusters that can isolate failure modes, giving you insights on how to address these failures with data augmentations.
For this post, I will use the MNIST handwritten digits dataset to demonstrate how embedding visualization works.
FiftyOne Zoo contains toy datasets for the purpose of demonstration. First, the MNIST dataset is downloaded from FiftyOne Zoo.
import fiftyone as fo
import fiftyone.zoo as foz
dataset = foz.load_zoo_dataset("mnist")
Split 'train' already downloaded
Split 'test' already downloaded
Loading 'mnist' split 'train'
100% |█████████████| 60000/60000 [17.7s elapsed, 0s remaining, 3.4K samples/s]
Loading 'mnist' split 'test'
100% |█████████████| 10000/10000 [3.0s elapsed, 0s remaining, 3.3K samples/s]
Dataset 'mnist' created
For illustration, only a subset of the dataset is needed. I will use the test subset, since it only contains 10,000 images.
test_split = dataset.match_tags("test")
test_split
Dataset: mnist
Media type: image
Num samples: 10000
Tags: ['test']
Sample fields:
id: fiftyone.core.fields.ObjectIdField
filepath: fiftyone.core.fields.StringField
tags: fiftyone.core.fields.ListField(fiftyone.core.fields.StringField)
metadata: fiftyone.core.fields.EmbeddedDocumentField(fiftyone.core.metadata.Metadata)
ground_truth: fiftyone.core.fields.EmbeddedDocumentField(fiftyone.core.labels.Classification)
View stages:
1. MatchTags(tags=['test'], bool=True)
Using match_tags makes getting data subsets quick and easy. Each entry in the subset shares the same test tag. These tags can be modified if the customized dataset comes from an image directory. Instead of a train-validation-test tag split, tag split with the class names is possible.
Next, embeddings for a low-dimensional representation of the data is calculated. Luckily, this does not have to be written: FiftyOne provides its own implementation.
import cv2
import numpy as np
import fiftyone.brain as fob
# Construct a ``num_samples x num_pixels`` array of images
embeddings = np.array([
cv2.imread(f, cv2.IMREAD_UNCHANGED).ravel()
for f in test_split.values("filepath")
])
# Compute 2D representation
results = fob.compute_visualization(
test_split,
embeddings=embeddings,
num_dims=2,
method="umap",
brain_key="mnist_test",
verbose=True,
seed=51,
)
Generating visualization...
UMAP(random_state=51, verbose=True)
Sat Apr 16 16:49:47 2022 Construct fuzzy simplicial set
Sat Apr 16 16:49:47 2022 Finding Nearest Neighbors
Sat Apr 16 16:49:47 2022 Building RP forest with 10 trees
Sat Apr 16 16:49:47 2022 NN descent for 13 iterations
1 / 13
2 / 13
3 / 13
4 / 13
Stopping threshold met -- exiting after 4 iterations
Sat Apr 16 16:49:53 2022 Finished Nearest Neighbor Search
Sat Apr 16 16:49:55 2022 Construct embedding
Epochs completed: 100% |-------------| 500/500 [00:10]
Sat Apr 16 16:50:06 2022 Finished embedding
There are a few algorithms that can be used to produce a low-dimensional representation. Well established ones are UMAP, t-SNE and PCA. As of this post’s writing, UMAP is considered to be the best algorithm owing to its superior speed, better understandable parameters and preservation of global structures. However, its performance can actually vary based on the input data. Understanding UMAP does a good job explaining the nuances of choosing UMAP over something like t-SNE.
Visualization
Before we look at the embeddings result, we can first take a peek at the dataset.
session = fo.launch_app(view=test_split)
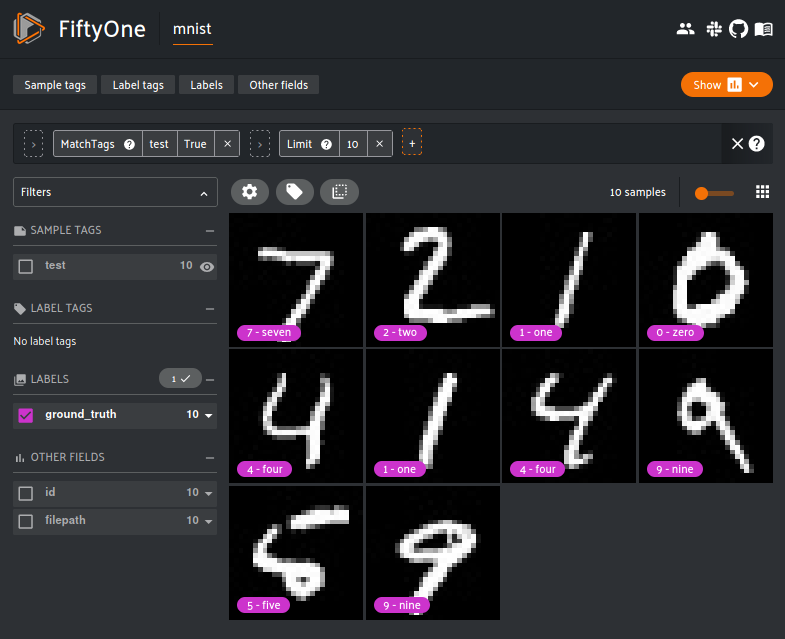
The FiftyOne App is a convenient GUI for browsing, visualizing and interacting directly with the dataset. All the user needs to do is supply the FiftyOne dataset object. The App can be used either locally, through the cloud or inside a Jupyter or Colab notebook.
The filters can be cascaded to get even finer subsets of the dataset. The above example chains the MatchTags and Limit filter in action. First, the MatchTags filter only allows data with the ‘test’ tags is displayed. The Limit filter allows the first 10 pictures to be displayed. With this, only the first 10 test data out of 10,000 data is shown. Creatively chaining the filters can produce some very fine grained data subset.
Once the dataset visualization is done, it is time to view the embeddings visualization results.
# Plot embeddings colored by ground truth label
plot = results.visualize(labels="ground_truth.label")
plot.show(height=720)
# Attach plot to session
session.plots.attach(plot)
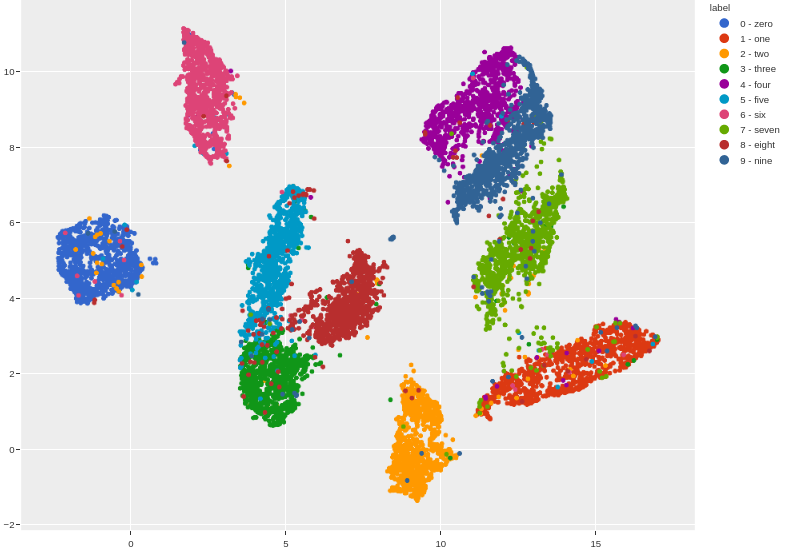
The separation of each class is quite distinct and can be seen even without outlining. The vast majority of the data are clustered correctly. However, each clusters also have outliers belonging to other clusters.
To take a look at these outliers relative to the cluster, the lasso tool can be used to isolate the points. For example, if I want to see the class 2 outliers in the class 0 cluster, I can first filter out the other classes, then lasso the outliers.
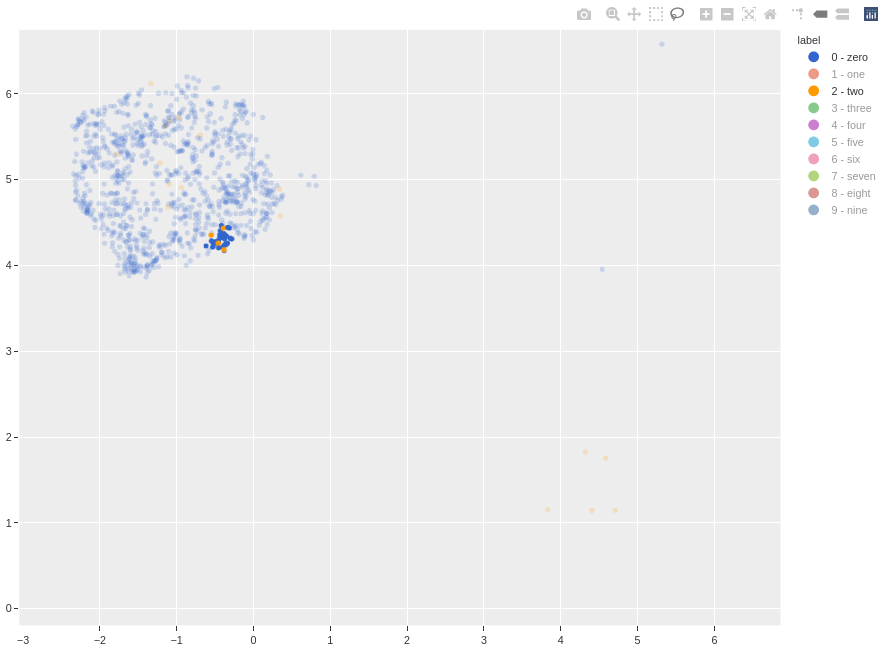
Resetting the FiftyOne App displays the lassoed points.
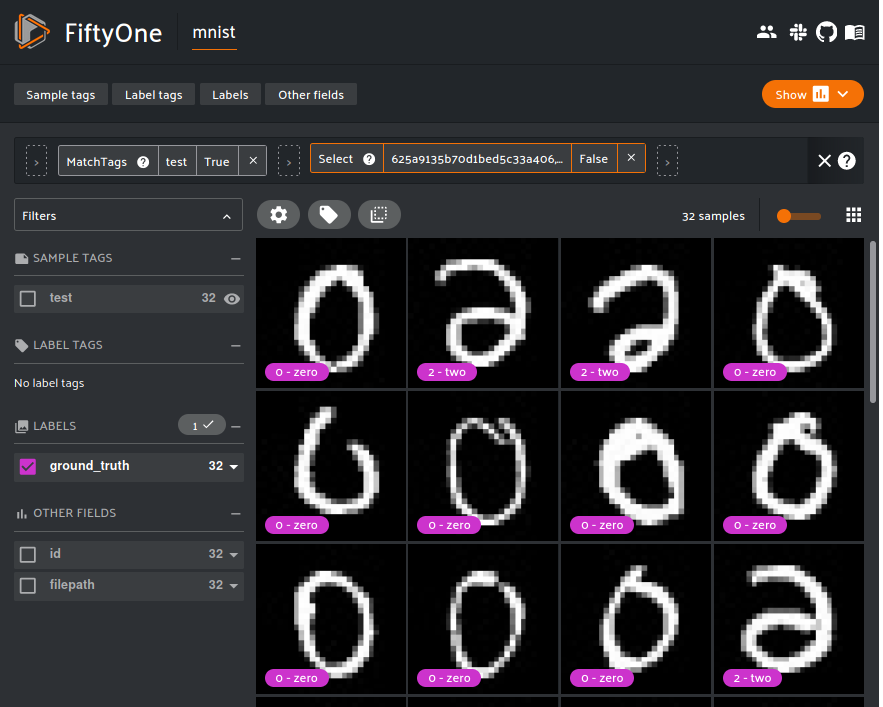
From the three class 2 outliers we can deduce the failure mode: some of the class 2 hand-writings are poor. If the tail is not written correctly, these 2s will look like either a reflected 6 or 0, which is probably why these data are incorrectly clustered as 0s.
With the findings there are some improvements that can be done. These outliers can either be re-annotated or removed from the dataset entirely. Given my previous points, removing these outliers is probably a safer bet.
Conclusion
We looked at the motivations of using specialized tools like FiftyOne to do data visualization. Specifically, we looked at how embeddings visualization can produce clusters that can reveal interesting insights, such as the data quality and failure modes. Explanations for outliers and suggestions to deal with them are briefly looked at.
Of course, FiftyOne comes with a lot of other powerful tools and features. Those who wish to improve their machine learning pipeline should really consider using FiftyOne or similar tools.
That’s all for this post. Thanks for reading, and stay tuned for future posts.